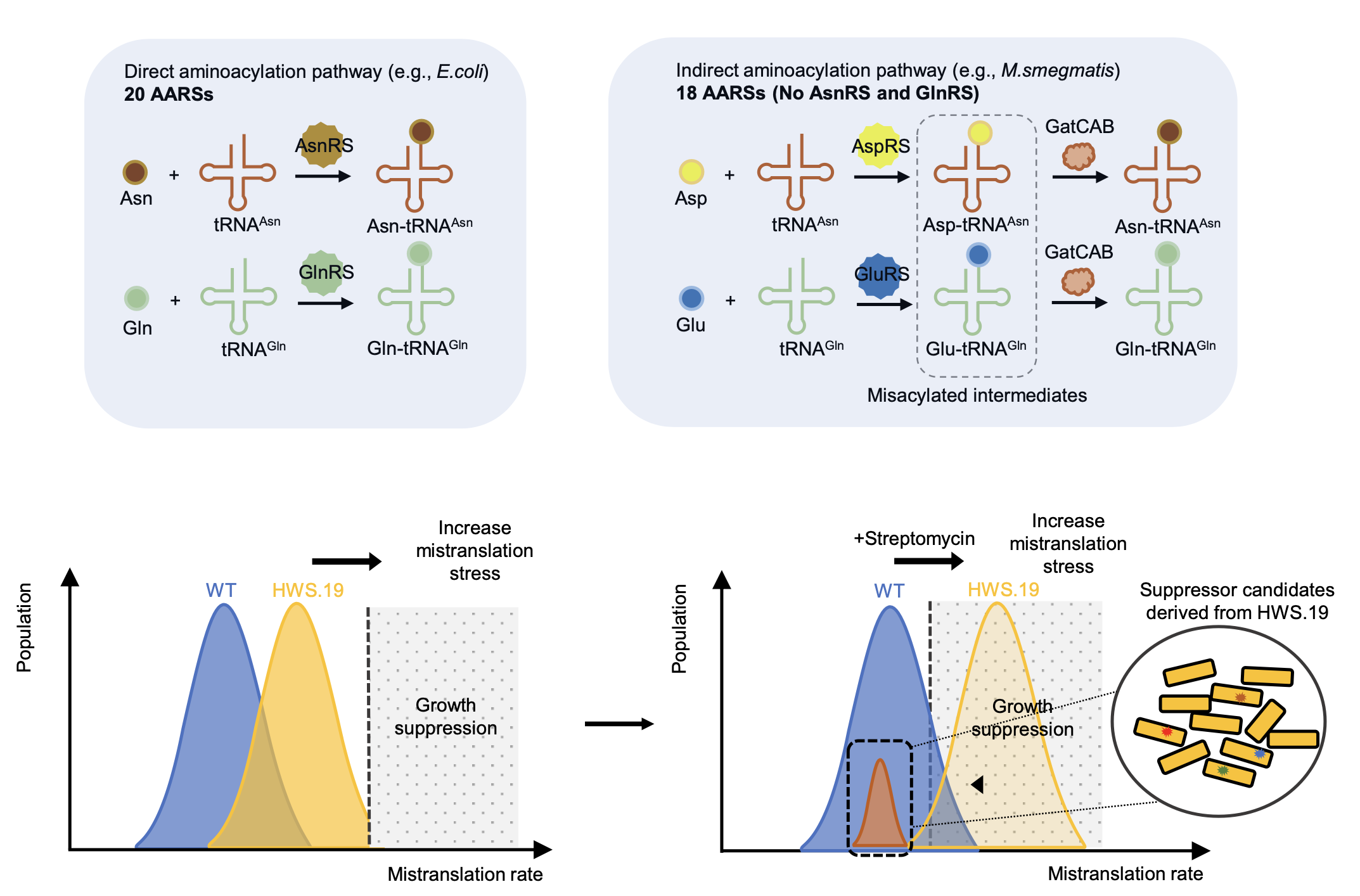
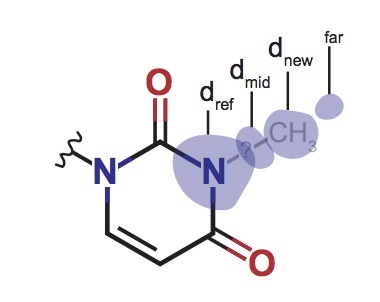
Ribosomal RNA methylation by GidB modulates discrimination of mischarged tRNA
Young ID*, Bi Z*, Chen YX*, Su HW, Chen Y, Hong JY, Fraser JS, Javid B.
bioRxiv,
2024
Structural characterization of ligand binding and pH-specific enzymatic activity of mouse Acidic Mammalian Chitinase
Díaz RE, Ecker AK, Correy GJ, Asthana P, Young ID, Faust B, Thompson MC, Seiple IB, Van Dyken SJ, Locksley RM, Fraser JS.
eLife,
2024
Access the Work
- Biorxiv Preprint: 542675
- Full Text
- Deposited Structures:
8FG5,
8FG7,
8GCA,
8FRC,
8FR9,
8FRB,
8FRD,
8FRG,
8FRA
- Zenodo Records:
8250616 (Kinetic properties of mAMCase catalytic domain at various pH),
7967930 (Characterization of mAMCase sugar-binding subsites),
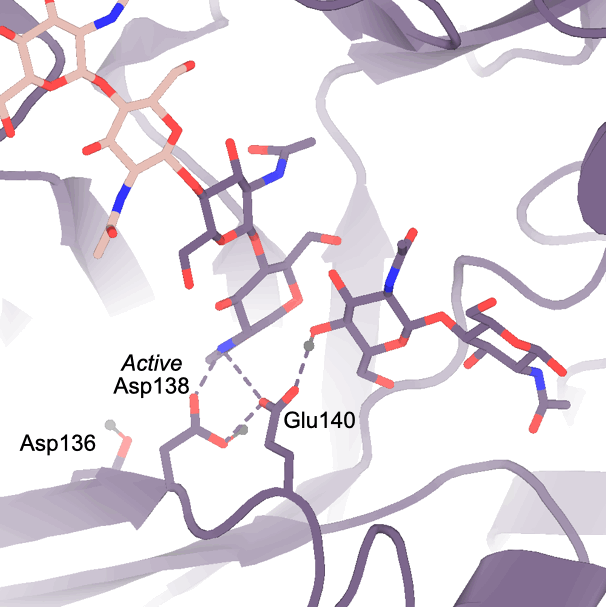
7905828 (Asp138 orientation correlates with ligand subsite occupancy),
7905863 (pKa of GH18 chitinases in the inactive and active conformation),
7758821 (10 ns Molecular Dynamics simulations of mAMCase at pH 2.0 and 6.5 in complex with GlcNAc6),
7967958 (Proposed catalytic mechanism of mAMCase),
7905944 (Crystallization conditions of apo and holo mAMCase),
7967978 (Overview of key residues for mAMCase activity),
7967954 (Protein-ligand interactions between mAMCase and chitin),
7758815 (Ringer analysis of catalytic triad confirms alternative Asp138 conformations)
Core Collaborators and Facilities
- Fraser lab @ UCSF
- Locksley lab @ UCSF
- Seiple lab @ UCSF
- Thompson lab @ UC Merced
- Van Dyken lab @ Washington University in St. Louis
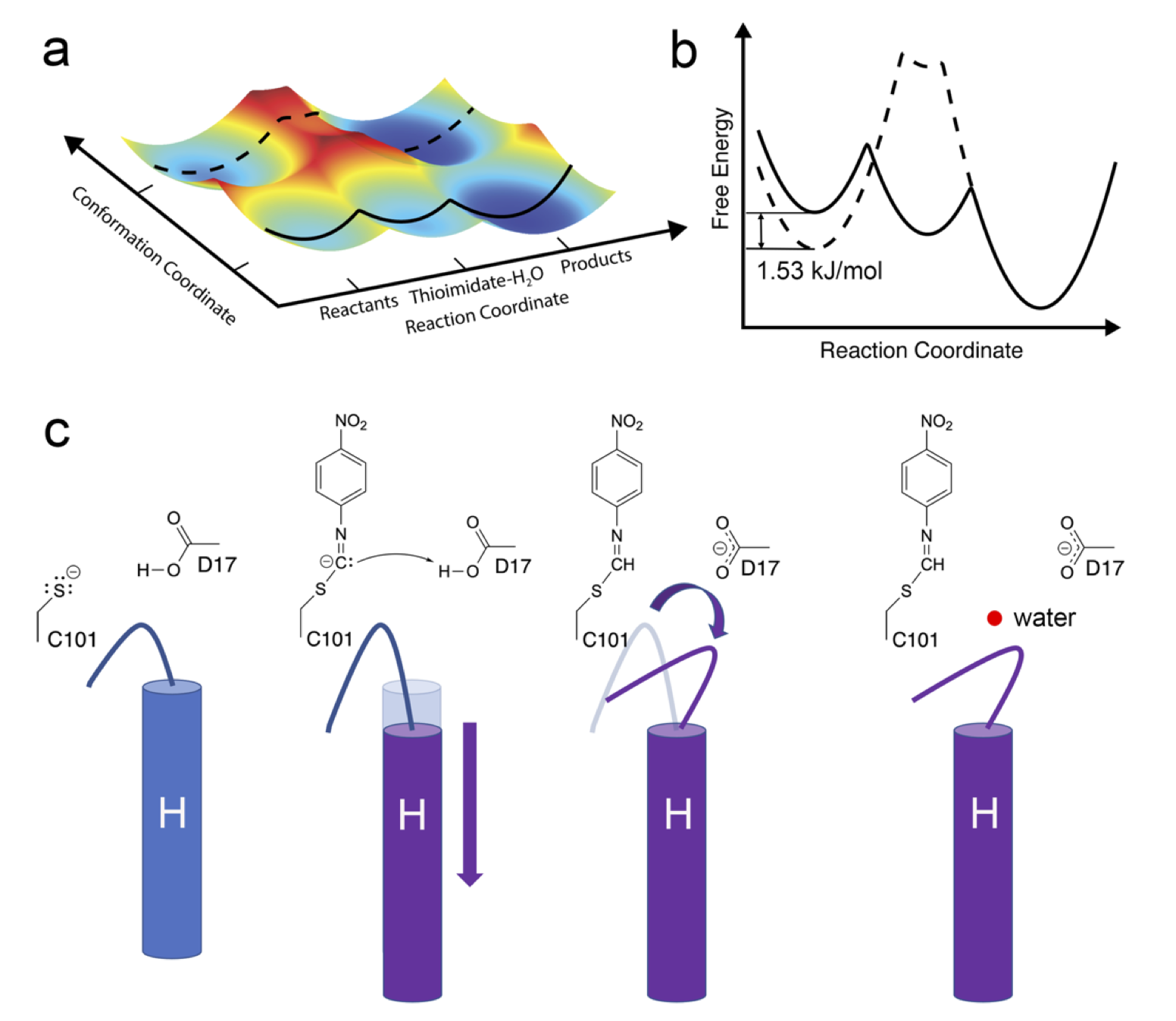
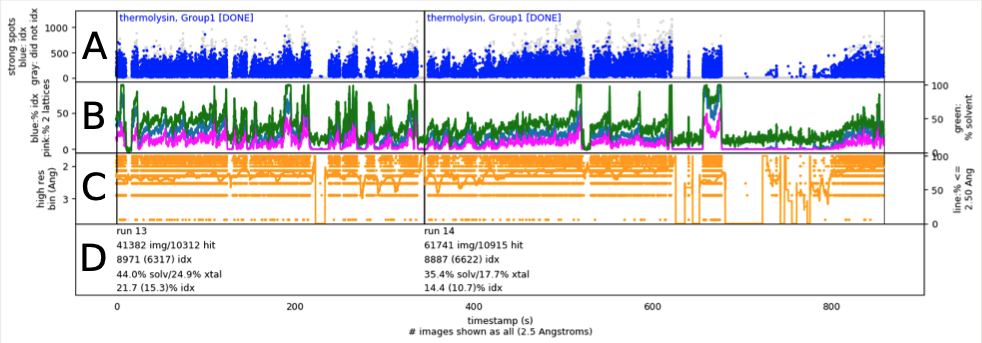
Changes in an enzyme ensemble during catalysis observed by high- resolution XFEL crystallography
Smith N, Dasgupta M, Wych DC, Dolamore C, Sierra RG, Lisova S, Marchany-Rivera D, Cohen AE, Boutet S, Hunter MS, Kupitz C, Poitevin F, Moss III FR, Mittan-Moreau DW, Brewster AS, Sauter NK, Young ID, Wolff AM, Tiwari VK, Kumar N, Berkowitz DB, Hadt RG, Thompson MC, Follmer AH, Wall ME, Wilson MA.
Science Advances,
2024
Access the Work
- PMID: 38536910
- PMCID: PMC10971408
- Biorxiv Preprint: 553460
- Full Text
- Deposited Structures:
8VPW,
8VQ1,
8TSX,
8TSU,
8TSY,
8TSZ,
8TT0,
8TT1,
8TT2,
8TT4,
8TT5
Core Collaborators and Facilities
- Hadt lab @ Caltech
- LCLS MFX endstation
- Sauter group @ LBNL
- Thompson lab @ UC Merced
- Wilson lab @ Nebraska
- Michael Wall @ LANL
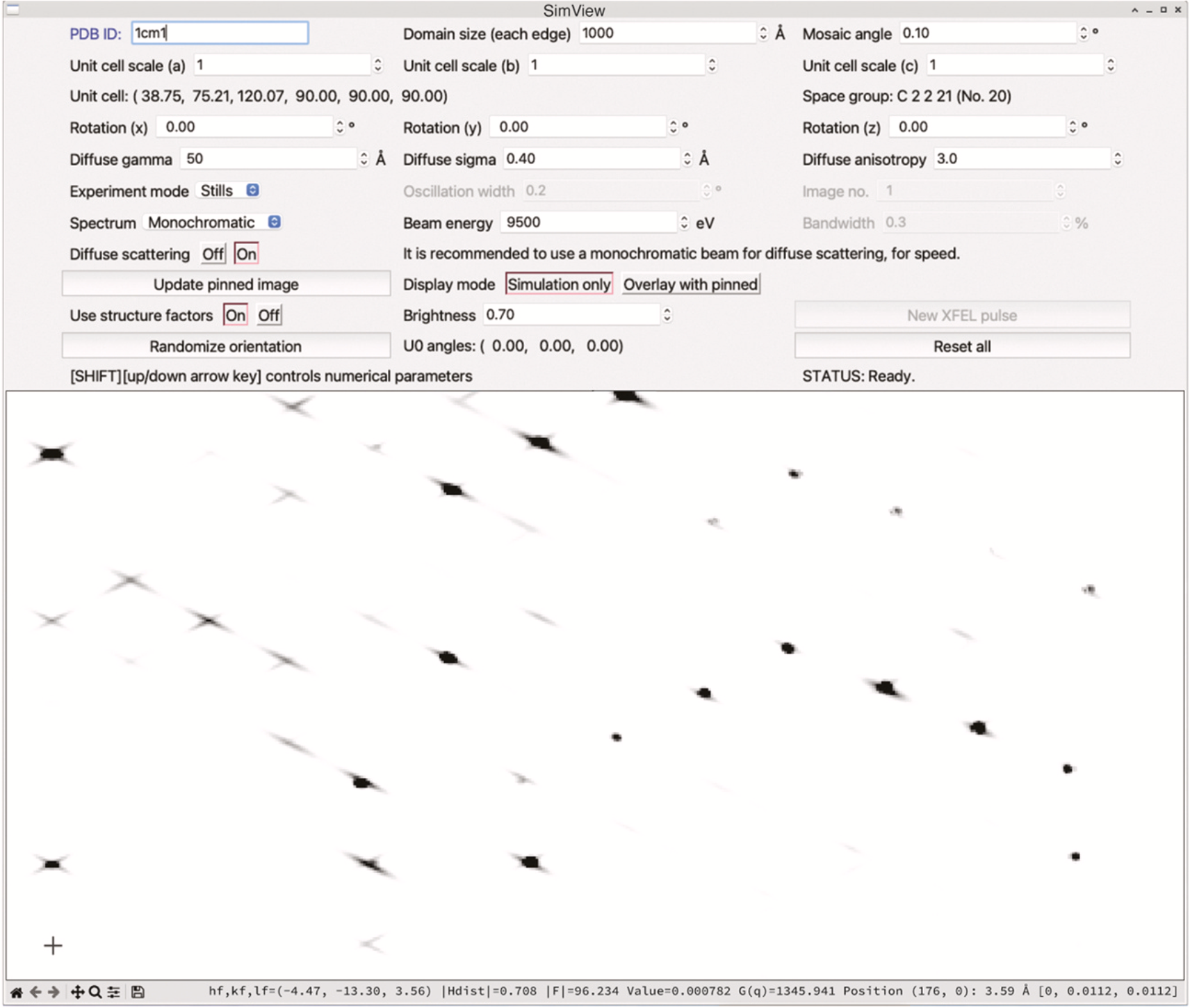
Interpreting macromolecular diffraction through simulation
Young ID, Mendez D, Poon BK, Blaschke JP, Wittwer F, Wall ME, Sauter NK
Methods in Enzymology,
2023
Mapping protein dynamics at high spatial resolution with temperature-jump X-ray crystallography
Wolff AM, Nango E, Young ID, Brewster AS, Kubo M, Nomura T, Sugahara M, Owada S, Barad BA, Ito K, Bhowmick A, Carbajo S, Hino T, Holton JM, Im D, O’Riordan LJ, Tanaka T, Tanaka R, Sierra RG, Yumoto F, Tono K, Iwata S, Sauter NK, Fraser JS, Thompson MC.
Nature Chemistry,
2023
Access the Work
- Biorxiv Preprint: 495662
- Full Text
- Deposited Structures:
8CVU,
8CVV,
8CVW,
8CW0,
8CW1,
8CW3,
8CW5,
8CW6,
8CW7,
8CW8,
8CWB,
8CWC,
8CWD,
8CWE,
8CWF,
8CWG,
8CWH
- GitHub Repository:
https://github.com/mthompson-lab/Temperature-Jump-Crystallography_Analysis-for-Paper (T-jump analysis code)
Additional Link
- T-jump raw diffraction images in CXIDB
Core Collaborators and Facilities
- Thompson lab @ UC Merced
- Fraser lab @ UCSF
- Sauter group @ LBNL
- SACLA XFEL
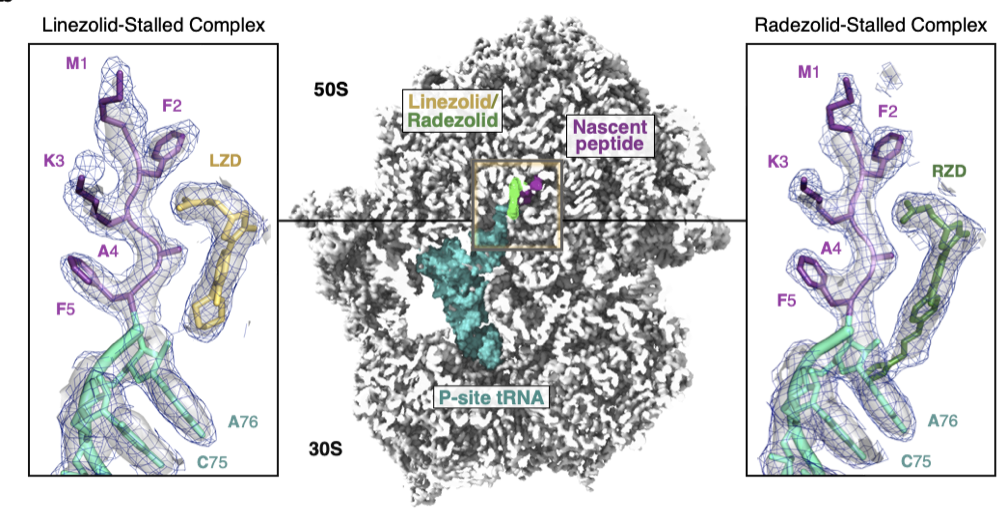
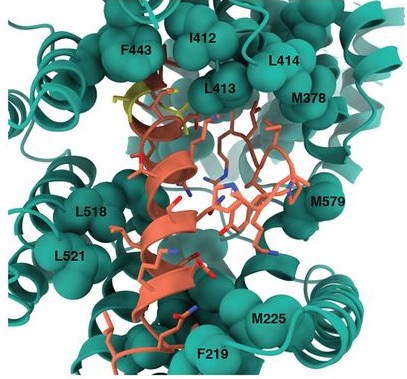
Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Tsai K*, Stojković V*, Lee DJ*, Young ID, Szal T, Vazquez-Laslop N, Mankin AS, Fraser JS, Fujimori DG.
Nature Structural and Molecular Biology,
2022
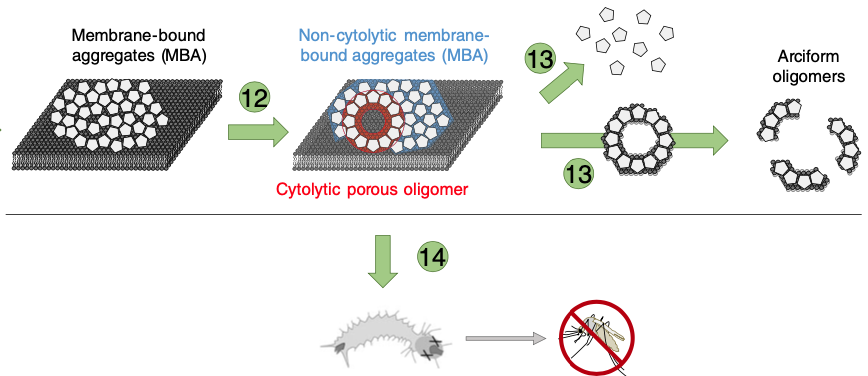
De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals
Tetreau G*, Sawaya MR*, De Zitter E*, Andreeva EA+, Banneville A-S+, Schibrowsky N+, Coquelle N, Brewster AS, Grünbein ML, Kovacs GN, Hunter MS, Kloos M, Sierra RG, Schiro G, Qiao P, Stricker M, Bideshi D, Young ID, Zala N, Engilberge S, Gorel A, Signor L, Teulon J-M, Hilpert M, Foucar L, Bielecki J, Bean R, de Wijn R, Sato T, Kirkwood H, Letrun R, Batyuk A, Snigireva I, Fenel D, Schubert R, Canfield EJ, Alba MM, Laporte F, Després L, Bacia M, Roux A, Chapelle C, Riobé F, Maury O, Ling WL, Boutet S, Mancuso A, Gutsche I, Girard E, Barends TRM, Pellequer J-L, Park H-W, Laganowsky AD, Rodriguez J, Burghammer M, Shoeman RL, Doak RB, Weik M, Sauter NK, Federici B, Cascio D, Schlichting I, Colletier J-P.
Nature Communications,
2022
Directed evolution of the rRNA methylating enzyme Cfr reveals molecular basis of antibiotic resistance.
Tsai K, Stojković V, Noda-Garcia L, Young ID, Myasnikov AG, Kleinman J, Palla A, Floor SN, Frost A, Fraser JS, Tawfik DS, Fujimori DG.
eLife,
2022
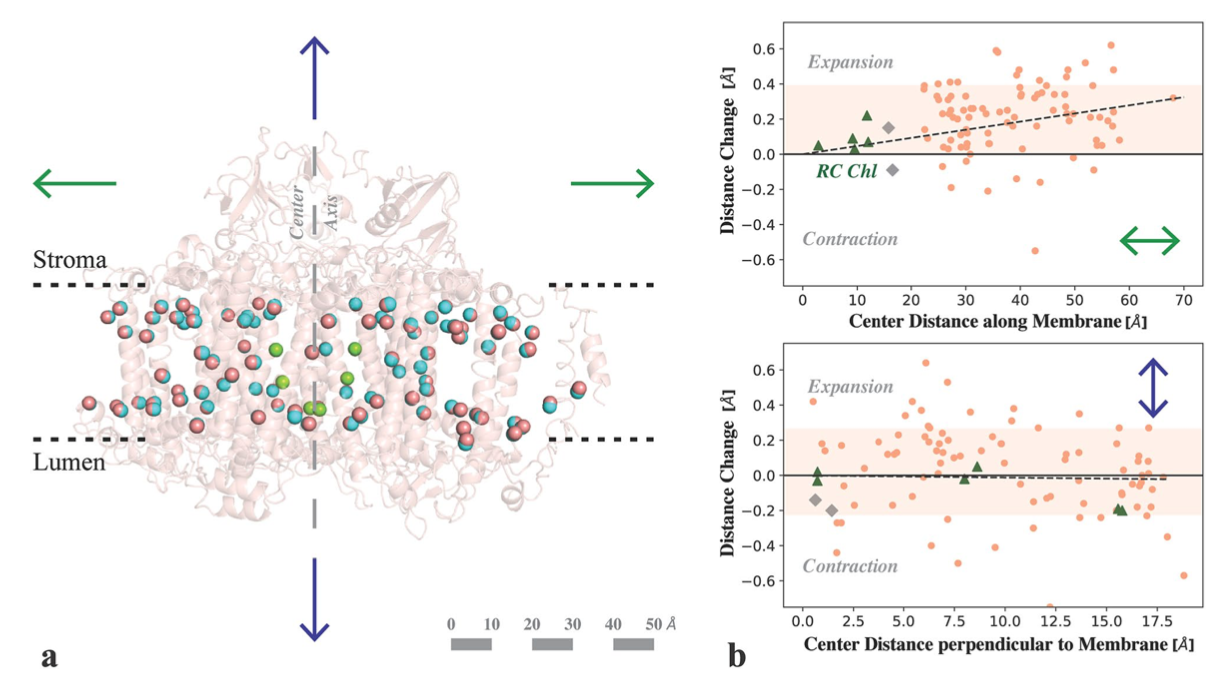
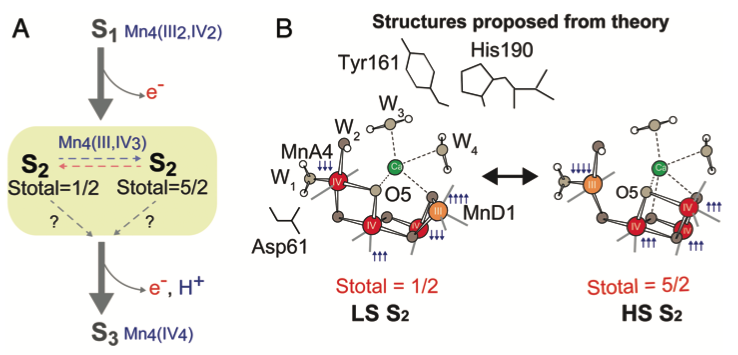
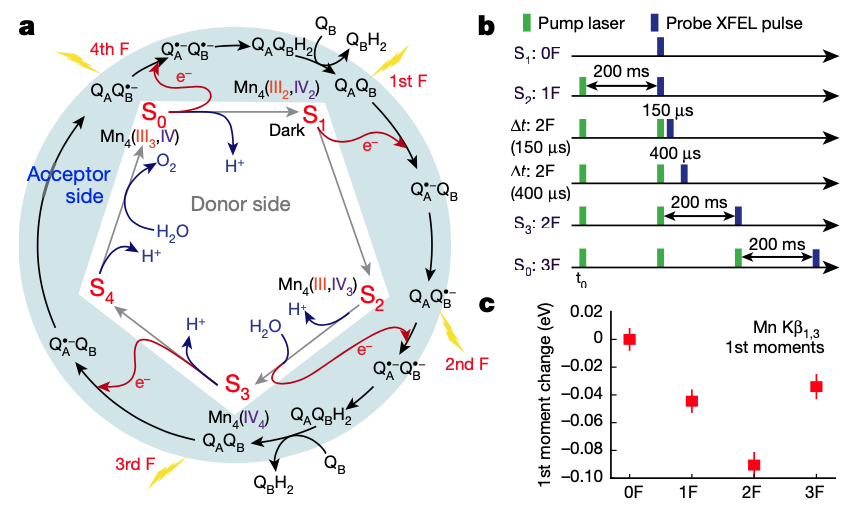
Structural dynamics in the water and proton channels of photosystem II during the S2 to S3 transition
Hussein R*, Ibrahim M*, Bhowmick A*, Simon PS*, Chatterjee R*, Lassalle L, Doyle M, Bogacz I, Kim I-S, Cheah MH, Gul S, de Lichtenberg C, Chernev P, Pham CC, Young ID, Carbajo S, Fuller FD, Alonso-Mori R, Batyuk A, Sutherlin KD, Brewster AS, Bolotovsky R, Mendez D, Holton JM, Moriarty NW, Adams PD, Bergmann U, Sauter NK, Dobbek H, Messinger J, Zouni A, Kern J, Yachandra VK, Yano J.
Nature Communications,
2021
Room temperature XFEL crystallography reveals asymmetry in the vicinity of the two phylloquinones in photosystem I
Keable SM*, Kölsch A*, Simon PS*, Dasgupta M, Chatterjee R, Subramanian SK, Hussein R, Ibrahim M, Kim I-S, Bogacz I, Makita H, Pham CC, Fuller FD, Gul S, Paley D, Lassalle L, Sutherlin KD, Bhowmick A, Moriarty NW, Young ID, Blaschke JP, de Lichtenberg C, Chernev P, Cheah MH, Park S, Park G, Kim J, Lee SJ, Park J, Tono K, Owada S, Hunter MS, Batyuk A, Oggenfuss R, Sander M, Zerdane S, Ozerov D, Nass K, Lemke H, Mankowsky R, Brewster AS, Messinger J, Sauter NK, Yachandra VK, Yano J, Zouni A, Kern J.
Scientific Reports,
2021
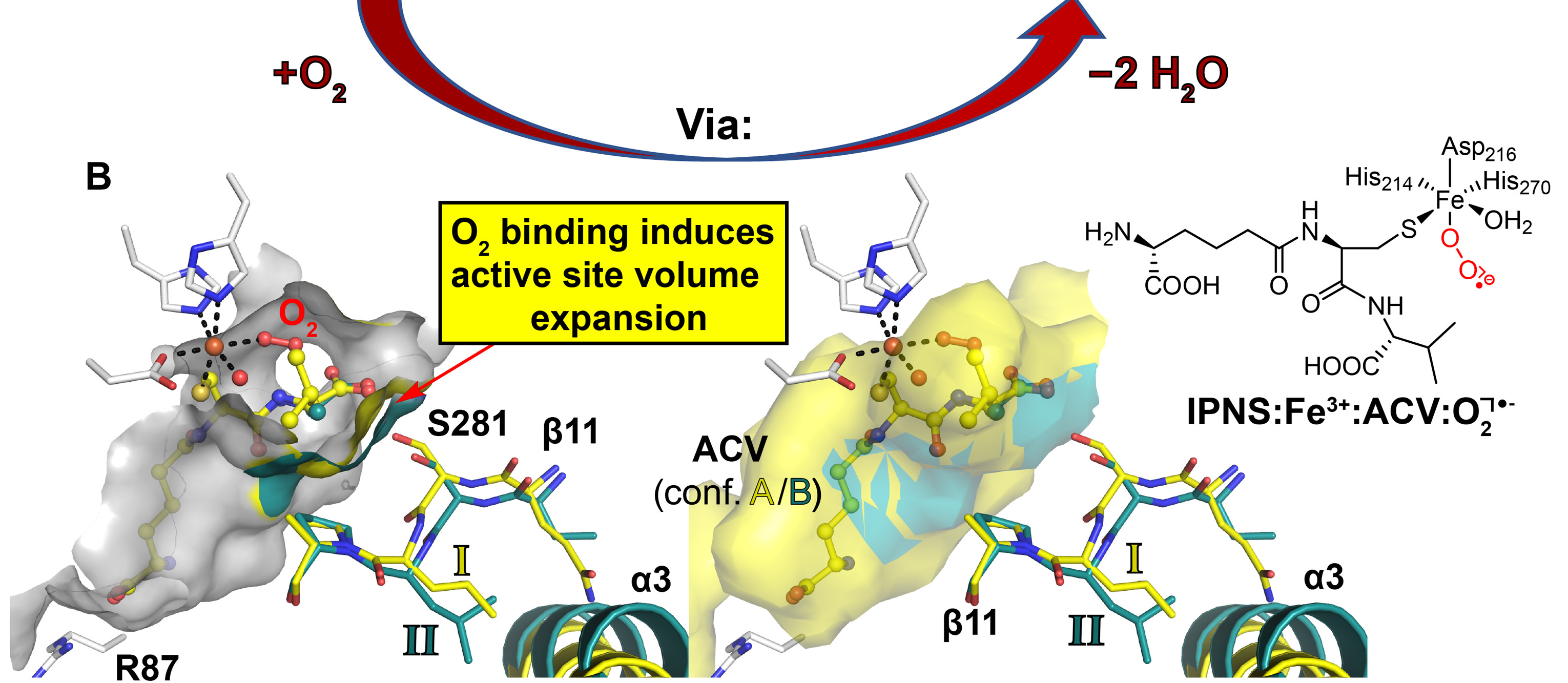
X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis
Rabe P, Kamps JJAG, Sutherlin KD, Linyard JDS, Aller P, Pham CC, Makita H, Clifton J, McDonough MA, Leissing TM, Shutin D, Lang PA, Butryn A, Brem J, Gul S, Fuller FD, Kim IS, Cheah MH, Fransson T, Bhowmick A, Young ID, O’Riordan L, Brewster AS, Pettinati I, Doyle M, Joti Y, Owada S, Tono K, Batyuk A, Hunter MS, Alonso-Mori R, Bergmann U, Owen RL, Sauter NK, Claridge TDW, Robinson CV, Yachandra VK, Yano J, Kern JF, Orville AM, Schofield CJ
Science Advances,
2021
Access the Work
- PMID: 34417180
- PMCID: PMC8378823
- Full Text
- Deposited Structures:
6ZAE,
6ZAF,
6ZAG,
6ZAH,
6ZAI,
6ZAJ,
6ZAL,
6ZAQ,
6Y0P,
6ZAO,
6ZAN,
6ZAP,
6ZAM,
6ZW8
Core Collaborators and Facilities
- Schofield lab @ Oxford
- Orville group @ Diamond Light Source
- Yano-Yachandra-Kern lab @ LBNL
- Robinson group @ Oxford
- Sauter group @ LBNL
- SACLA XFEL
- Diamond Light Source
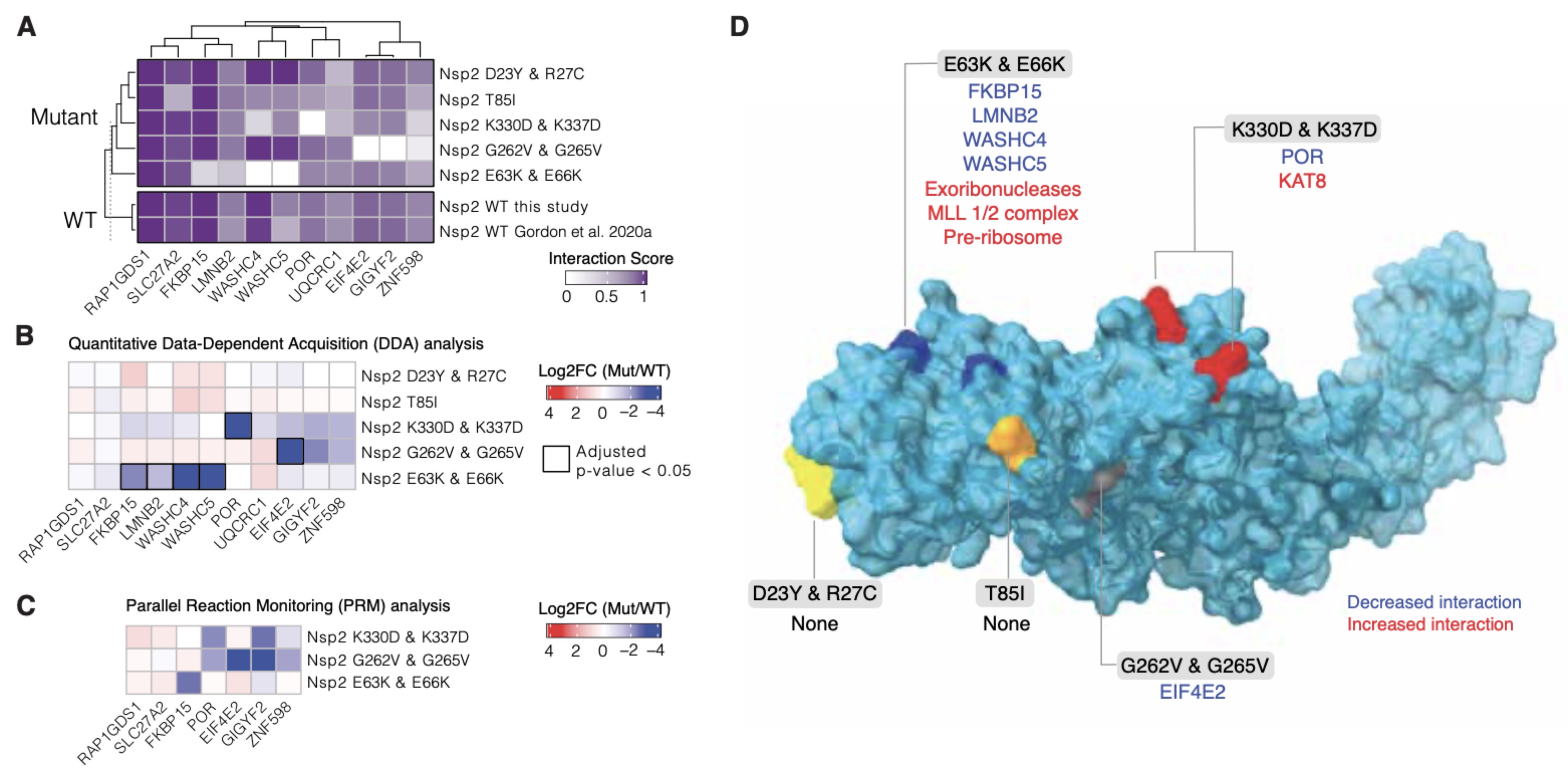
CryoEM and AI reveal a structure of SARS-CoV-2 Nsp2, a multifunctional protein involved in key host processes.
Gupta M*, Azumaya CM*, Moritz M*, Pourmal S*, Diallo A*, Merz GE*, Jang G*, Bouhaddou M*, Fossati*, Brilot AF, Diwanji D, Hernandez E, Herrera N, Kratochvil HT, Lam VL, Li F, Li Y, Nguyen HC, Nowotny C, Owens TW, Peters JK, Rizo AN, Schulze-Gahmen U, Smith AM, Young ID, Yu Z, Asarnow D, Billesbølle C, Campbell MG, Chen J, Chen KH, Chio US, Dickinson MS, Doan L, Jin M,, Kim K, Li J, Li YL, Linossi E, Liu Y, Lo M, Lopez J, Lopez KE, Mancino A, Moss III FR, Paul MD, Pawar KI, Pelin A, Pospiech Jr. TH, Puchase C, Remesh SG, Safari M, Schaefer K, Sun M, Tabios MC, Thwin AC, Titus EW, Trenker R, Tse E, Tsui TKM, Wang F, Zhang K, Zhang Y, Zhao J, Zhou F, Zhou Y, Zuliani-Alvarez L, QCRG Structural Biology Consortium, Agard DA, Cheng Y, Fraser JS, Jura N, Kortemme T, Manglik A, Southworth DR, Stroud RM, Swaney DL, Krogan NJ, Frost A, Rosenberg OS, Verba KA.
Research Square (preprint under review),
2021
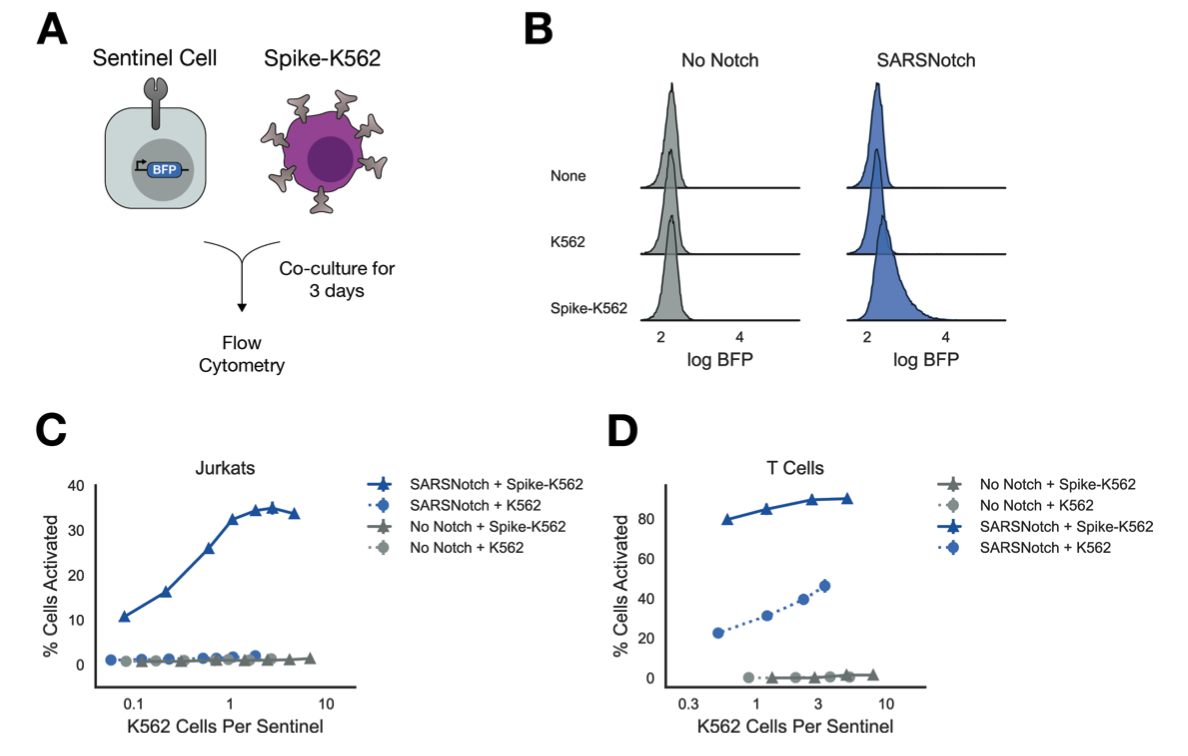
Sentinel cells enable genetic detection of SARS-CoV-2 Spike protein
Weinberg ZY, Hilburger CE, Kim M, Cao L, Khalid M, Elmes S, Diwanji D, Hernandez E, Lopez J, Schaefer K, Smith AM, Zhou F, QCRG Structural Biology Consortium, Kumar GR, Ott M, Baker D, El-Samad H.
bioRxiv,
2021
Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Schuller M*, Correy GJ*, Gahbauer S*, Fearon D*, Wu T, Díaz RE, Young ID, Carvalho Martins L, Smith DS, Schulze-Gahmen U, Owens TW, Deshpande I, Merz GE, Thwin AC, Biel JT, Peters JK, Mortiz M, Herrera N, Kratochvil HT, QCRG Structural Biology Consortium, Aimon A, Bennett MJ, Brandao Neto J, Cohen EA, Dias A, Douangamath A, Dunnett L, Fedorov O, Ferla PM, Fuchs M, Gorrie-Stone JT, Holton MJ, Johnson GM, Krojer T, Meigs G, Powell JA, Rack J, Rangel LV, Russi S, Skyner ER, Smith AC, Soares SA, Wierman LJ, Zhu K, O’Brien P, Jura N, Ashworth A, Irwin J, Thompson MC, Gestwicki JE, von Delft F, Shoichet BK, Fraser JS, Ahel I.
Science Advances,
2021
Access the Work
- PMID: 33853786
- PMCID: PMC8046379
- Full Text
- Deposited Structures:
7KR0,
7KR1,
7KQW,
7KQO,
7KQP,
5RVJ,
5RVK,
5RVL,
5RVM,
5RVN,
5RVO,
5RVP,
5RVQ,
5RVR,
5RVS,
5RVT,
5RVU,
5RVV,
5RS7,
5RS8,
5RS9,
5RSB,
5RSC,
5RSD,
5RSE,
5RSF,
5RSG,
5RSH,
5RSI,
5RSJ,
5RSK,
5RSL,
5RSM,
5RSN,
5RSO,
5RSP,
5RSQ,
5RSR,
5RSS,
5RST,
5RSU,
5RSV,
5RSW,
5RSX,
5RSY,
5RSZ,
5RT0,
5RT1,
5RT2,
5RT3,
5RT4,
5RT5,
5RT6,
5RT7,
5RT8,
5RT9,
5RTA,
5RTB,
5RTC,
5RTD,
5RTE,
5RTF,
5RTG,
5RTH,
5RTI,
5RTJ,
5RTK,
5RTL,
5RTM,
5RTN,
5RTO,
5RTP,
5RTQ,
5RTR,
5RTS,
5RTT,
5RTU,
5RTV,
5RTW,
5RTX,
5RTY,
5RTZ,
5RU0,
5RU1,
5RU2,
5RU3,
5RU4,
5RU5,
5RU6,
5RU7,
5RU8,
5RU9,
5RUA,
5RUC,
5RUD,
5RUE,
5RUF,
5RUG,
5RUH,
5RUI,
5RUJ,
5RUK,
5RUL,
5RUM,
5RUN,
5RUO,
5RUP,
5RUQ,
5RUR,
5RUS,
5RUT,
5RUU,
5RUV,
5RUW,
5RUX,
5RUY,
5RUZ,
5RV0,
5RV1,
5RV2,
5RV3,
5RV4,
5RV5,
5RV6,
5RV7,
5RV8,
5RV9,
5RVA,
5RVB,
5RVC,
5RVD,
5RVE,
5RVF,
5RVG,
5RVH,
5RVI,
5S6W,
5S18,
5S1A,
5S1C,
5S1E,
5S1G,
5S1I,
5S1K,
5S1M,
5S1O,
5S1Q,
5S1S,
5S1U,
5S1W,
5S1Y,
5S20,
5S22,
5S24,
5S26,
5S27,
5S28,
5S29,
5S2A,
5S2B,
5S2C,
5S2D,
5S2E,
5S2F,
5S2G,
5S2H,
5S2I,
5S2J,
5S2K,
5S2L,
5S2M,
5S2N,
5S2O,
5S2P,
5S2Q,
5S2R,
5S2S,
5S2T,
5S2U,
5S2V,
5S2W,
5S2X,
5S2Y,
5S2Z,
5S30,
5S31,
5S32,
5S33,
5S34,
5S35,
5S36,
5S37,
5S38,
5S39,
5S3A,
5S3B,
5S3C,
5S3D,
5S3E,
5S3F,
5S3G,
5S3H,
5S3I,
5S3J,
5S3K,
5S3L,
5S3M,
5S3N,
5S3O,
5S3P,
5S3Q,
5S3R,
5S3S,
5S3T,
5S3U,
5S3V,
5S3W,
5S3X,
5S3Y,
5S3Z,
5S40,
5S41,
5S42,
5S43,
5S44,
5S45,
5S46,
5S47,
5S48,
5S49,
5S4A,
5S4B,
5S4C,
5S4D,
5S4E,
5S4F,
5S4G,
5S4H,
5S4I,
5S4J,
5S4K
- Zenodo Records:
4716363 (UCSF C2 crystal form screen),
4716089 (UCSF P43 crystal form screen),
4606901 (Docking Fragment Screen),
4628855 (Differential Scanning Fluorimetry),
4628875 (Isothermal Titration Calorimetry)
- Addgene Plasmids:
169209 (C2 crystal form),
169210 (P43 crystal form)
Additional Links
- Original post
- Fraganalysis from Diamond Lightsource
- Celebratory Tweetstorm from James Fraser
- Celebratory Tweetstorm from Mike Thompson
- UCSF Press Release - Building Blocks for COVID-19 Antiviral Drugs Identified in Rapid Study
- Practical Fragments - Hundreds of fragments hits for the SARS-CoV-2 Nsp3 Macrodomain
- Massive fragment screen points way to new SARS-CoV-2 inhibitors
Core Collaborators and Facilities
- Ahel lab @ Oxford
- Fraser lab @ UCSF
- Diamond Light Source
- Advanced Light Source @ LBNL
- QBI Coronavirus Research Group @ UCSF
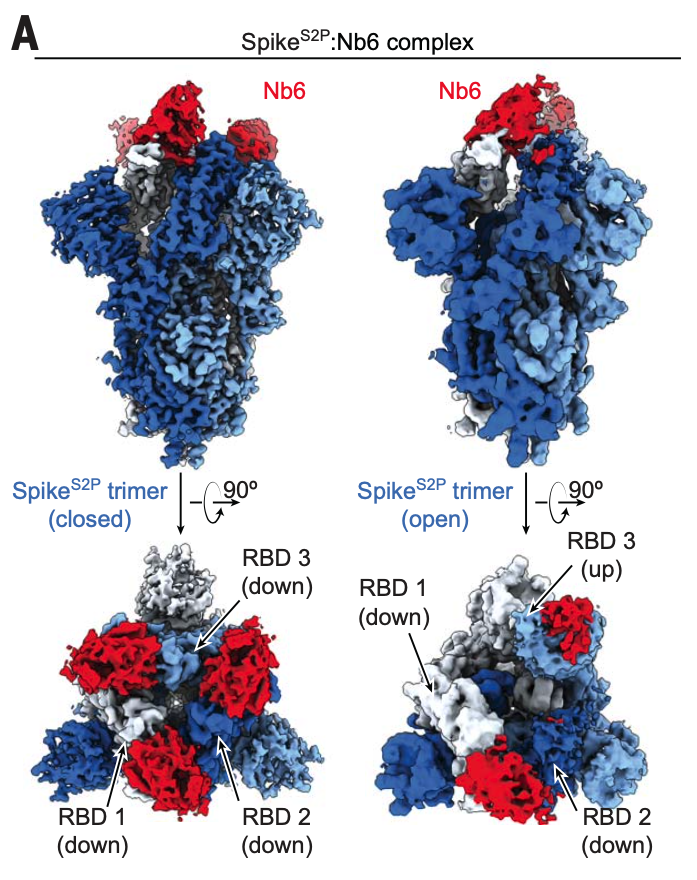
An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike
Schoof M*, Faust B*, Saunders RA*, Sangwan S*, Rezelj V*, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Deshpande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, García-Sastre A, Ott M, Vignuzzi M, QCRG Structural Biology Consortium, Walter P, Manglik A.
Science,
2020
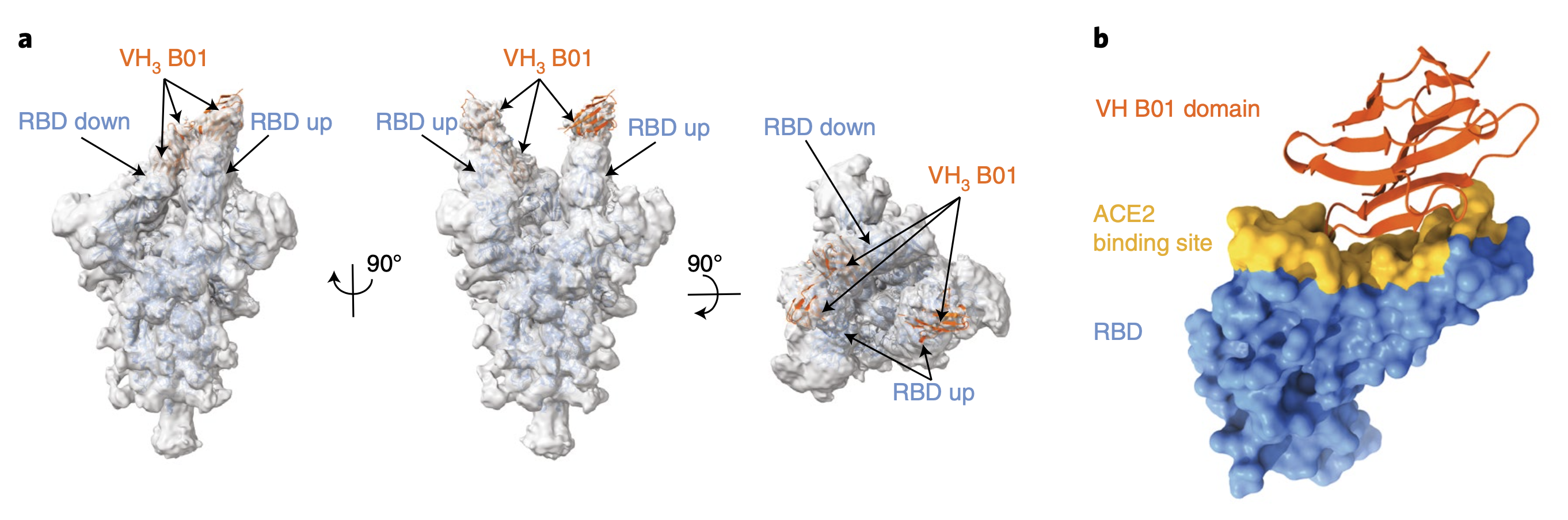
Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2.
Bracken CJ, Lim SA, Solomon P, Rettko NJ, Nguyen DP, Zha BS, Schaefer K, Byrnes JR, Zhou J, Lui I, Liu J, Pance K, QCRG Structural Biology Consortium, Zhou XX, Leung KK, Wells JA
Nature Chemical Biology,
2020
Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms.
Gordon DE*, Hiatt J*, Bouhaddou M*, Rezelj VV*, Ulferts S*, Braberg H*, Jureka AS*, Obernier K*, Guo JZ*, Batra J*, Kaake RM*, Weckstein AR*, Owens TW*, Gupta M*, Pourmal S*, Titus EW*, Cakir M*, Soucheray M, McGregor M, Cakir Z, Jang G, O’Meara MJ, Tummino TA, Zhang Z, Foussard H, Rojc A, Zhou Y, Kuchenov D, Hüttenhain R, Xu J, Eckhardt M, Swaney DL, Fabius JM, Ummadi M, Tutuncuoglu B, Rathore U, Modak M, Haas P, Haas KM, Naing ZZC, Pulido EH, Shi Y, Barrio-Hernandez I, Memon D, Petsalaki E, Dunham A, Marrero MC, Burke D, Koh C, Vallet T, Silvas JA, Azumaya CM, Billesbølle C, Brilot AF, Campbell MG, Diallo A, Dickinson MS, Diwanji D, Herrera N, Hoppe N, Kratochvil HT, Liu Y, Merz GE, Moritz M, Nguyen HC, Nowotny C, Puchades C, Rizo AN, Schulze-Gahmen U, Smith AM, Sun M, Young ID, Zhao J, Asarnow D, Biel J, Bowen A, Braxton JR, Chen J, Chio CM, Chio US, Deshpande I, Doan L, Faust B, Flores S, Jin M, Kim K, Lam VL, Li F, Li J, Li YL, Li Y, Liu X, Lo M, Lopez KE, Melo AA, Moss FR 3rd, Nguyen P, Paulino J, Pawar KI, Peters JK, Pospiech TH Jr, Safari M, Sangwan S, Schaefer K, Thomas PV, Thwin AC, Trenker R, Tse E, Tsui TKM, Wang F, Whitis N, Yu Z, Zhang K, Zhang Y, Zhou F, Saltzberg D, QCRG Structural Biology Consortium, Hodder AJ, Shun-Shion AS, Williams DM, White KM, Rosales R, Kehrer T, Miorin L, Moreno E, Patel AH, Rihn S, Khalid MM, Vallejo-Gracia A, Fozouni P, Simoneau CR, Roth TL, Wu D, Karim MA, Ghoussaini M, Dunham I, Berardi F, Weigang S, Chazal M, Park J, Logue J, McGrath M, Weston S, Haupt R, Hastie CJ, Elliott M, Brown F, Burness KA, Reid E, Dorward M, Johnson C, Wilkinson SG, Geyer A, Giesel DM, Baillie C, Raggett S, Leech H, Toth R, Goodman N, Keough KC, Lind AL; Zoonomia Consortium, Klesh RJ, Hemphill KR, Carlson-Stevermer J, Oki J, Holden K, Maures T, Pollard KS, Sali A, Agard DA, Cheng Y, Fraser JS, Frost A, Jura N, Kortemme T, Manglik A, Southworth DR, Stroud RM, Alessi DR, Davies P, Frieman MB, Ideker T, Abate C, Jouvenet N, Kochs G, Shoichet B, Ott M, Palmarini M, Shokat KM, García-Sastre A, Rassen JA, Grosse R, Rosenberg OS, Verba KA, Basler CF, Vignuzzi M, Peden AA, Beltrao P, Krogan NJ.
Science,
2020
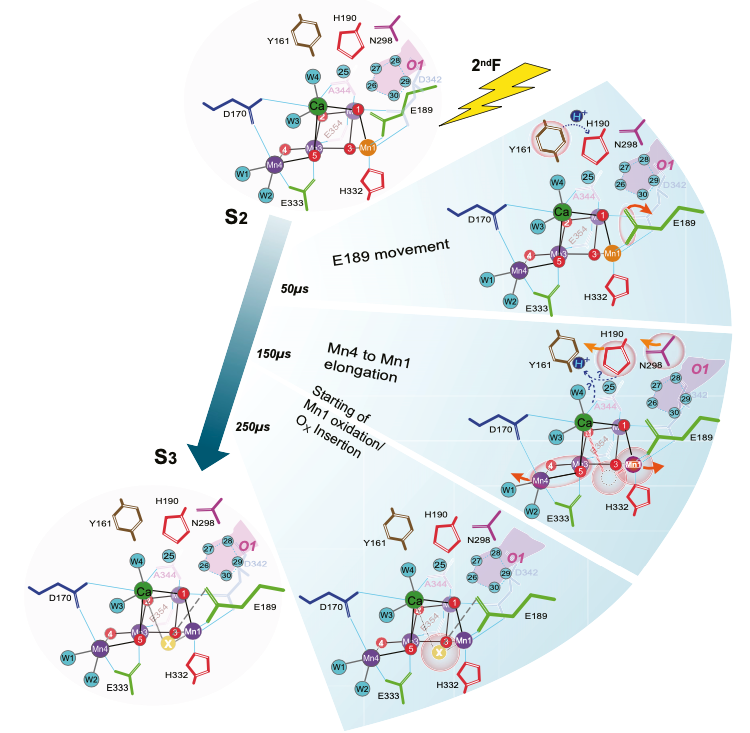
Untangling the sequence of events during the S2 → S3 transition in photosystem II and implications for the water oxidation mechanism
Ibrahim M*, Fransson T*, Chatterjee R*, Cheah MH*, Hussein R, Lassalle L, Sutherlin KD, Young ID, Fuller FD, Gul S, Kim I-S, Simon PS, de Lichtenberg C, Chernev P, Bogacz I, Pham CC, Orville AM, Saichek N, Northern T, Batyuk A, Carbajo S, Alonso-Mori R, Tono K, Owada S, Bhowmick A, Bolotovsky R, Mendez D, Moriarty NW, Holton JM, Dobbek H, Brewster AS, Adams PD, Sauter NK, Bergmann U, Zouni A, Messinger J, Kern J, Yachandra VK, Yano J.
Proceedings of the National Academy of Science,
2020
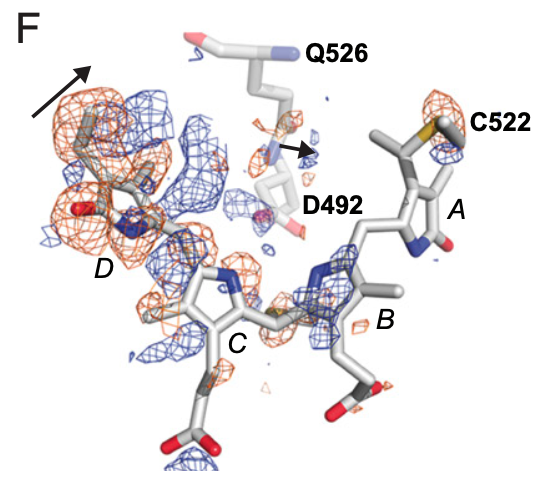
Assessment of the nucleotide modifications in the high-resolution cryo-electron microscopy structure of the Escherichia coli 50S subunit.
Stojković V, Myasnikov AG, Young ID, Frost A, Fraser JS, Fujimori DG.
Nucleic Acids Research,
2020
Serial femtosecond crystallography on in vivo-grown crystals drives elucidation of mosquitocidal Cyt1Aa bioactivation cascade
Tetreau G*, Banneville A-S*, Andreeva EA*, Brewster AS*, Hunter MS, Sierra RG, Teulon J-M, Young ID, Burke N, Grünewald TA, Beaudouin J, Snigireva I, Fernandez-Luna MT, Burt A, Park H-W, Signor L, Bafna JA, Sadir R, Fenel D, Boeri-Erba E, Bacia M, Zala N, Laporte F, Després L, Weik M, Boutet S, Rosenthal M, Coquelle N, Burghammer M, Cascio D, Sawaka MR, Winterhalter M, Gratton E, Gutsche I, Federici B, Pellequer J-L, Sauter NK, Colletier J-P.
Nature Communications,
2020
Comparing serial X-ray crystallography and microcrystal electron diffraction (MicroED) as methods for routine structure determination from small macromolecular crystals.
Wolff AM, Young ID, Sierra RG, Brewster AS, Martynowycz MW, Nango E, Sugahara M, Nakane T, Ito K, Aquila A, Bhowmick A, Biel JT, Carbajo S, Cohen AE, Cortez S, Gonzalez A, Hino T, Im D, Koralek JD, Kubo M, Lazarou TS, Nomura T, Owada S, Samelson A, Tanaka R, Tanaka T, Thompson EM, van den Bedem H, Woldeyes RA, Yumoto F, Zhao W, Tono K, Boutet S, Iwata S, Gonen T, Sauter NK, Fraser JS, Thompson MC.
IUCrJ,
2020
Photoreversible interconversion of a phytochrome photosensory module in the crystalline state
Burgie ES*, Clinger JA*, Miller MD, Brewster AS, Aller P, Butryn A, Fuller FD, Gul S, Young ID, Pham CC, Kim I-S, Bhowmick A, O’Riordan LJ, Sutherlin KD, Heinemann JV, Batyuk A, Alonso-Mori R, Hunter MS, Koglin JE, Yano J, Yachandra VK, Sauter NK, Cohen AE, Kern J, Orville AM, Phillips Jr GN, Vierstra RD.
Proceedings of the National Academy of Science,
2020
Structural isomers of the S2 state in photosystem II: do they exist at room temperature and are they important for function?
Chatterjee R, Lassalle L, Gul S, Fuller FD, Young ID, Ibrahim M, de Lichtenberg C, Cheah MH, Zouni A, Messinger J, Yachandra VK, Kern J, Yano J.
Physiologia Plantarum,
2019
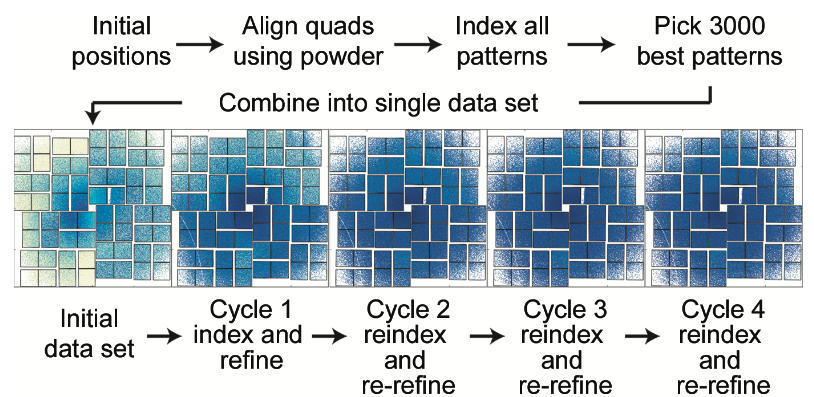
Processing serial crystallographic data from XFELs or synchrotrons using the cctbx.xfel GUI
Brewster AS, Young ID, Lyubimov AY, Bhowmick A, Sauter NK.
Computational Crystallographic Newsletter,
2019
Structures of the intermediates of Kok’s photosynthetic water oxidation clock
Kern J, Chatterjee R*, Young ID*, Fuller FD*, Lassalle L, Ibrahim M, Gul S, Fransson T, Brewster AS, Alonso-Mori R, Hussein R, Zhang M, Douthit L, de Lichtenberg C, Cheah MH, Shevela D, Wersig J, Seuffert I, Sokaras D, Pastor E, Weninger C, Kroll T, Sierra RG, Aller P, Butryn A, Orville AM, Liang M, Batyuk A, Koglin JE, Carbajo S, Boutet S, Moriarty NW, Holton JM, Dobbek H, Adams PD, Bergmann U, Sauter NK, Zouni A, Messinger J, Yano J, Yachandra VK.
Nature,
2018
Improving signal strength in serial crystallography with DIALS geometry refinement
Brewster AS, Waterman DG, Parkhurst JM, Gildea RJ, Young ID, O’Riordan LJ, Yano J, Winter G, Evans G, Sauter NK.
Acta Crystallographica D: Structural Biology,
2018
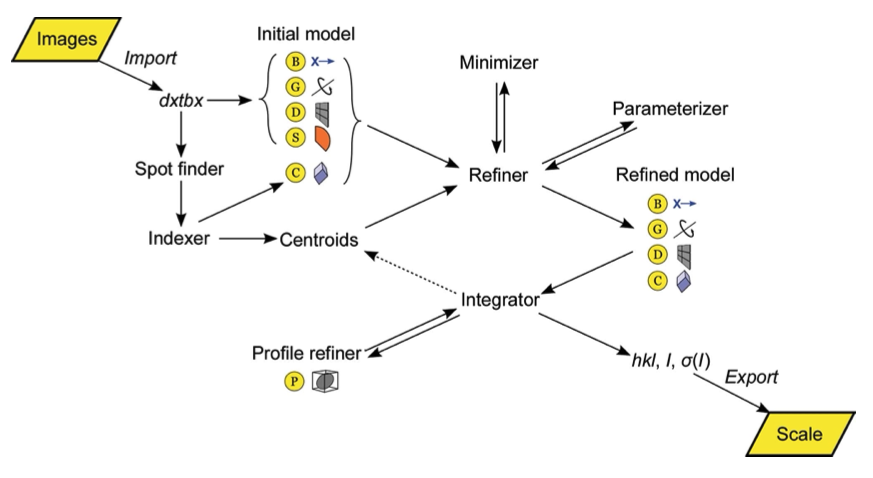
DIALS: implementation and evaluation of a new integration package
Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G.
Acta Crystallographica D: Structural Biology,
2018
High-speed fixed-target serial virus crystallography
Roedig P*, Ginn HM*, Pakendorf T, Sutton G, Harlos K, Walter TS, Meyer J, Fischer P, Duman R, Vartiainen I, Reime B, Warmer M, Brewster AS, Young ID, Michels-Clark T, Sauter NK, Kotecha A, Kelly J, Rowlands DJ, Sikorsky M, Nelson S, Damiani DS, Alonso-Mori R, Ren J, Fry EE, David C, Stuart DI, Wagner A, Meents A.
Nature Methods,
2017
High-speed fixed-target serial virus crystallography
Meents A, Young ID, Wagner AH, Stuart DI, David C, Fry EE, Ren J, Alonso-Mori R, Damiani DS, Nelson S, Sikorsky M, Sauter NK, Michels Clark T, Roedig P, Brewster AS, Warmer M, Reime B, Vartiainen I, Duman R, Fischer P, Meyer J, Walter TS, Harlos K, Sutton G, Pakendorf T, Ginn HM.
Protocol Exchange,
2017
Drop-on-demand sample delivery for studying biocatalysts in action at X-ray free-electron lasers
Fuller FD*, Gul S*, Chatterjee R, Bergie ES, Young ID, Lebrette H, Srinivas V, Brewster AS, Michels-Clark T, Clinger JA, Andi B, Ibrahim M, Pastor E, de Lichtenberg C, Hussein R, Pollock CJ, Zhang M, Stan CA, Kroll T, Fransson T, Weninger C, Kubin M, Aller P, Lassalle L, Bräuer P, Miller MD, Amin M, Koroidov S, Roessler CG, Allaire M, Sierra RG, Docker PT, Glownia JM, Nelson S, Koglin JE, Zhu D, Chollet M, Song S, Lemke H, Mengning L, Sokaras D, Alonso-Mori R, Zouni A, Messinger J, Bergmann U, Boal AK, Bollinger Jr JM, Krebs C, Högbom M, Phillips Jr GN, Vierstra RD, Sauter NK, Orville AM, Kern J, Yachandra VK, Yano J.
Nature Methods,
2017
Towards characterization of photo-excited electron transfer and catalysis in natural and artificial systems using XFELs
Alonso-Mori R, Asa K, Bergmann U, Brewster AS, Chatterjee R, Cooper JK, Frei HM, Fuller FD, Goggins E, Gul S, Fukuzawa H, Iablonskyi D, Ibrahim M, Katayama T, Kroll T, Kumagai Y, McClure BA, Messinger J, Motomura K, Nagaya K, Nishiyama T, Saracini C, Sato Y, Sauter NK, Sokaras D, Takanashi T, Togashi T, Ueda K, Weare WW, Weng T-C, Yabashi M, Yachandra VK, Young ID, Zouni A, Kern JF, Yano J.
Faraday Discussions,
2016
Structure of photosystem II and substrate binding at room temperature
Young ID*, Ibrahim M*, Chatterjee R*, Gul S, Fuller F, Koroidov S, Brewster AS, Tran R, Alonso-Mori R, Kroll T, Michels-Clark T, Laksmono H, Sierra RG, Stan CA, Hussein R, Zhang M, Douthit L, Kubin M, de Lichtenberg C, Vo PL, Nilsson H, Cheah MH, Shevela D, Saracini C, Bean MA, Seuffert I, Sokaras D, Weng T-C, Pastor E, Weninger C, Fransson T, Lassalle L, Bräuer P, Aller P, Docker PT, Andi B, Orville AM, Glownia JM, Nelson S, Sikorski M, Zhu D, Hunter MS, Lane TJ, Aquila A, Koglin JE, Robinson J, Liang M, Boutet S, Lyubimov AY, Uervirojnangkoorn M, Moriarty NW, Liebschner D, Afonine PV, Waterman DG, Evans G, Wernet P, Dobbek H, Weis WI, Brunger AT, Zwart PH, Adams PD, Zouni A, Messinger J, Bergmann U, Sauter NK, Kern J, Yachandra VK, Yano J.
Nature,
2016
Structural changes correlated with magnetic spin state isomorphism in the S2 state of the Mn4CaO5 cluster in the oxygen-evolving complex of photosystem II
Chatterjee R*, Han G*, Kern J, Gul S, Fuller FD, Garatchenko A, Young ID, Weng T-C, Norlund D, Alonso-Mori R, Bergmann U, Sokaras D, Hatakeyama M, Yachandra VK, Yano J.
Chemical Science,
2016
Processing XFEL data with cctbx.xfel and DIALS
Brewster AS, Waterman DG, Parkhurst JM, Gildea RJ, Michels-Clark TM, Young ID, Bernstein HJ, Winter G, Evans G, Sauter NK.
Computational Crystallographic Newsletter,
2016
Concentric-flow electrokinetic injector enables serial crystallography of ribosome and photosystem II
Sierra RG, Gati C, Laksmono H, Dao EH, Gul S, Fuller F, Kern J, Chatterjee R, Ibrahim M, Brewster AS, Young ID, Michels-Clark T, Aquila A, Liang M, Hunter MS, Koglin JE, Boutet S, Junco EA, Hayes B, Bogan MJ, Hampton CY, Puglisi EV, Sauter NK, Stan CA, Zouni A, Yano J, Yachandra VK, Soltis SM, Puglisi JD, DeMirci H.
Nature Methods,
2015