Peer Review
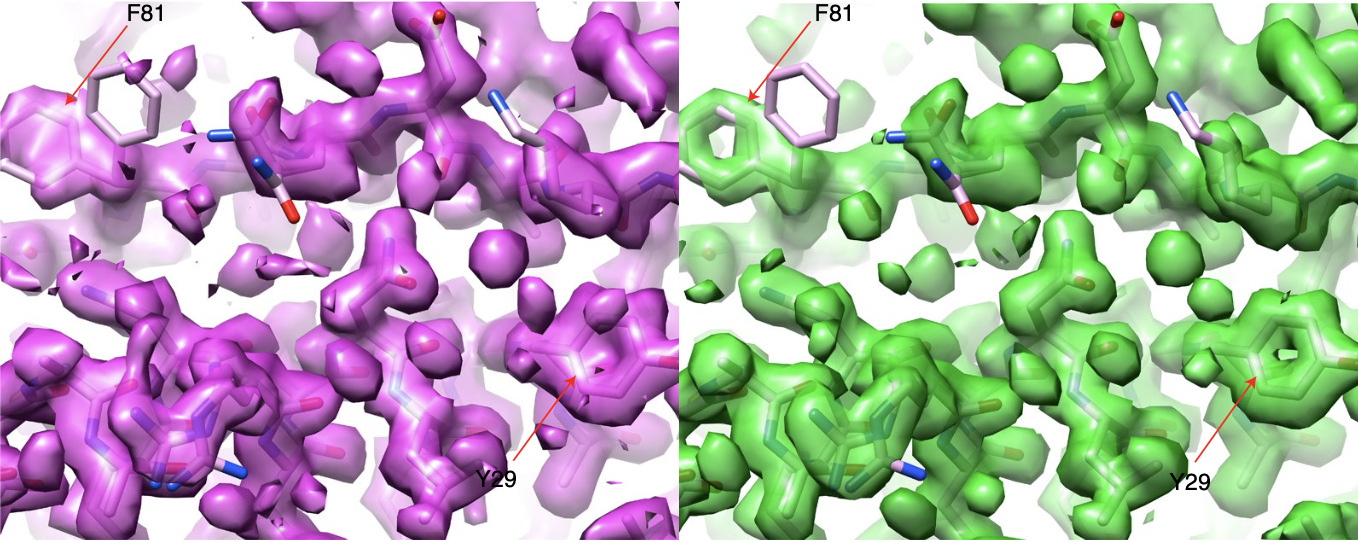
Ab initio phasing macromolecular structures using electron-counted MicroED data.
Martynowycz MW, Clabbers MTB, Hattne J, and Gonen T.
Reviewed by: Young ID, Fraser JS.
Effects of cryo-EM cooling on structural ensembles.
Bock LV, and Grubmüller H.
Reviewed by: Young ID, Fraser JS.
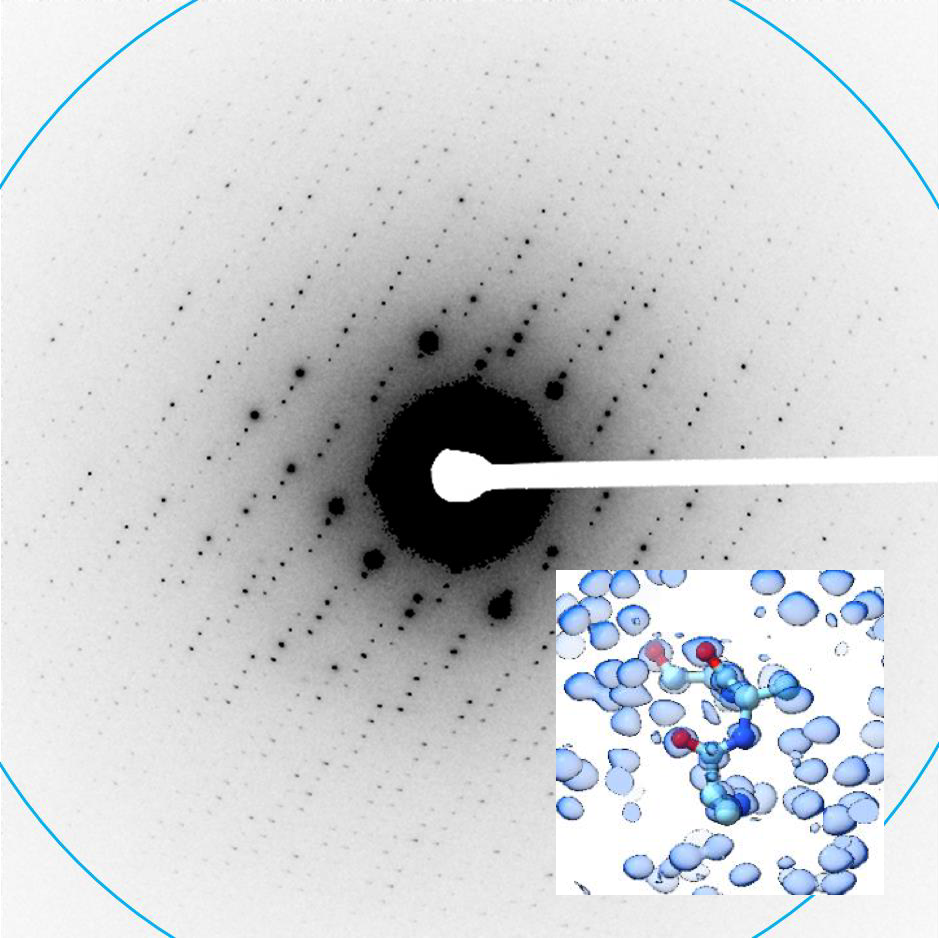
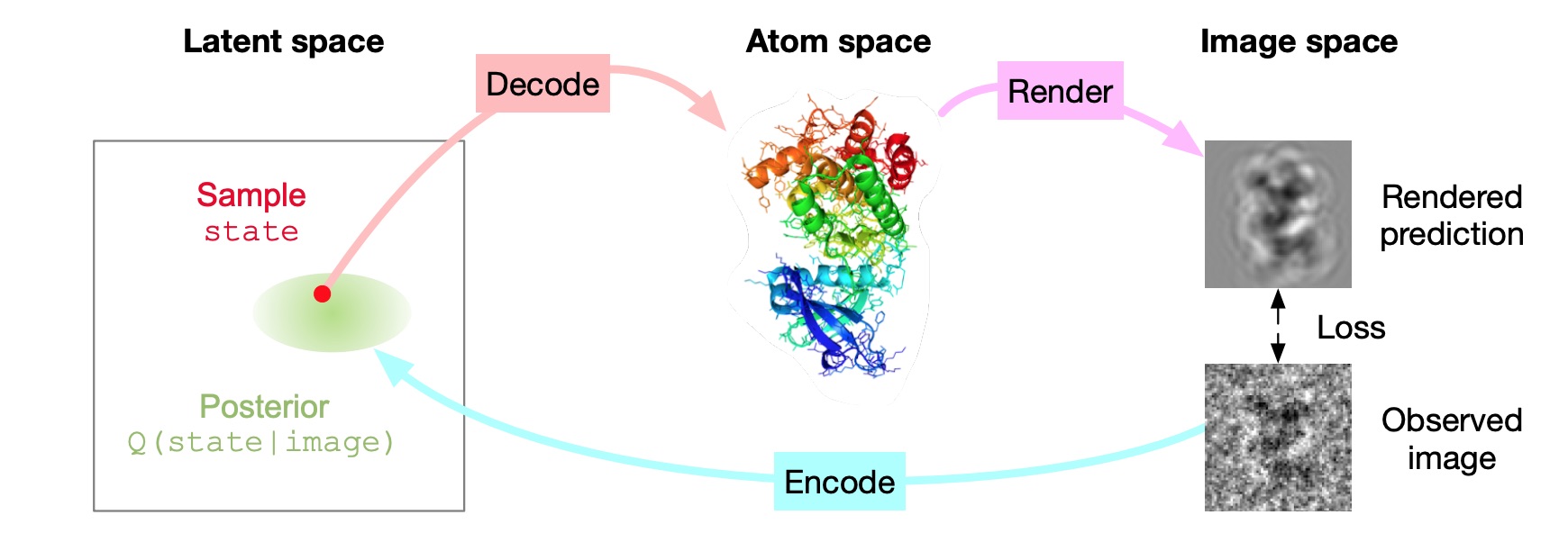
Inferring a Continuous Distribution of Atom Coordinates from Cryo-EM Images using VAEs.
Rosenbaum D, Garnelo M, Zielinski M, Beattie C, Clancy E, Huber A, Kohli P, Senior AW, Jumper J, Doersch C, Eslami SMA, Ronneberger O, and Adler J.
Reviewed by: Wankowicz SA, Young ID, Asarnow D, Fraser JS.
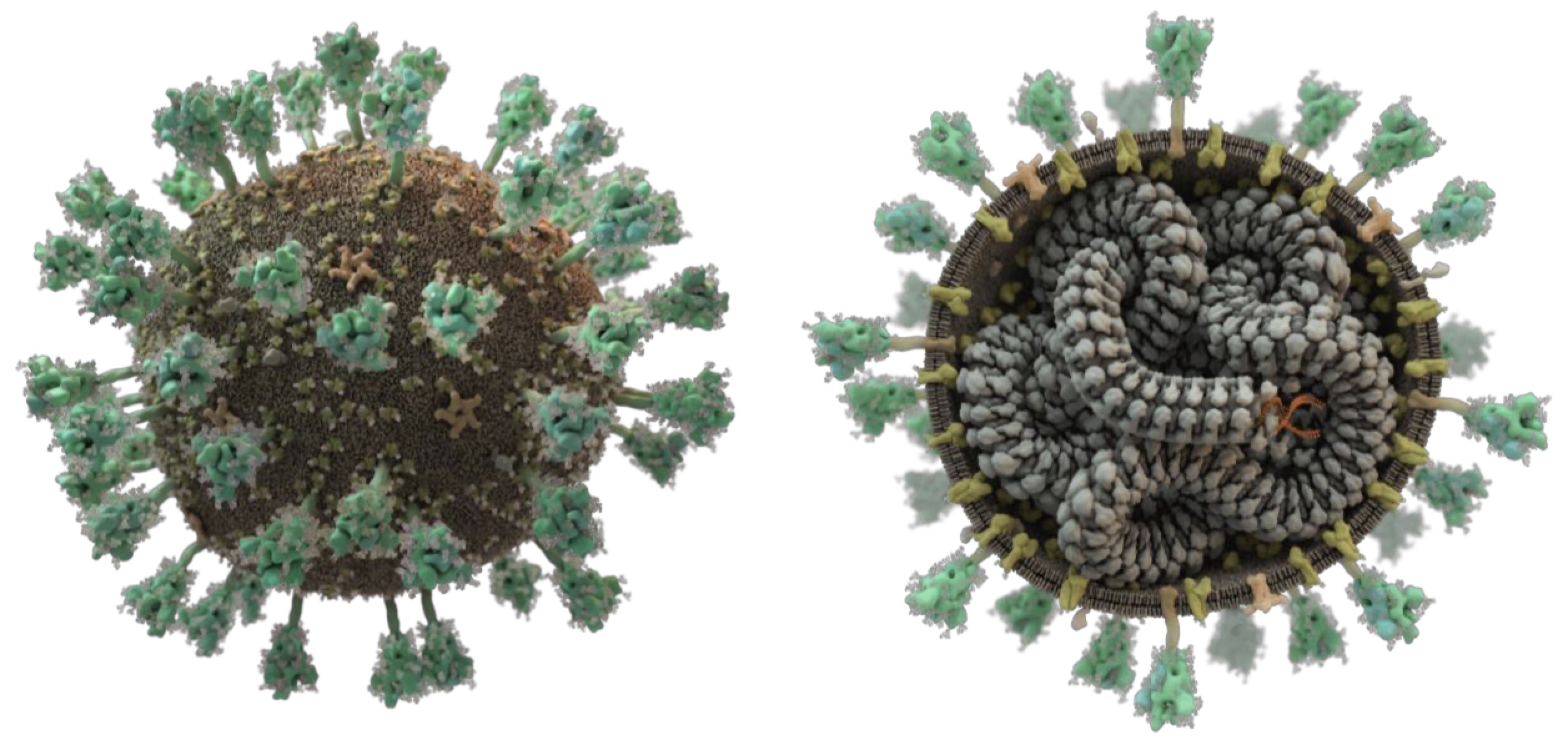
Improving SARS-CoV-2 structures: Peer review by early coordinate release.
Croll TI, Williams CJ, Chen VB, Richardson DC, and Richardson JS.
Reviewed by: Young ID, Fraser JS.
Croll and coworkers describe a unique form of peer-review enabled by the early release of biomolecular structure coordinates and density maps and detail its critical role in the search for SARS-CoV-2 vaccines and treatments.

- Zenodo Record: 4697901
Peer Review
reciprocalspaceship: a Python library for crystallographic data analysis.
Greisman JB, Dalton KM, and Hekstra DR.
Reviewed by: Young ID.
Making the invisible enemy visible.
Croll T, Diederichs K, Fischer F, Fyfe C, Gao Y, Horrell S, Joseph AP, Kandler L, Kippes O, Kirsten F, Müller K, Nolte K, Payne A, Reeves MG, Richardson J, Santoni G, Stäb S, Tronrud D, Williams C, and Thorn A.
Reviewed by: Young ID, Fraser JS.
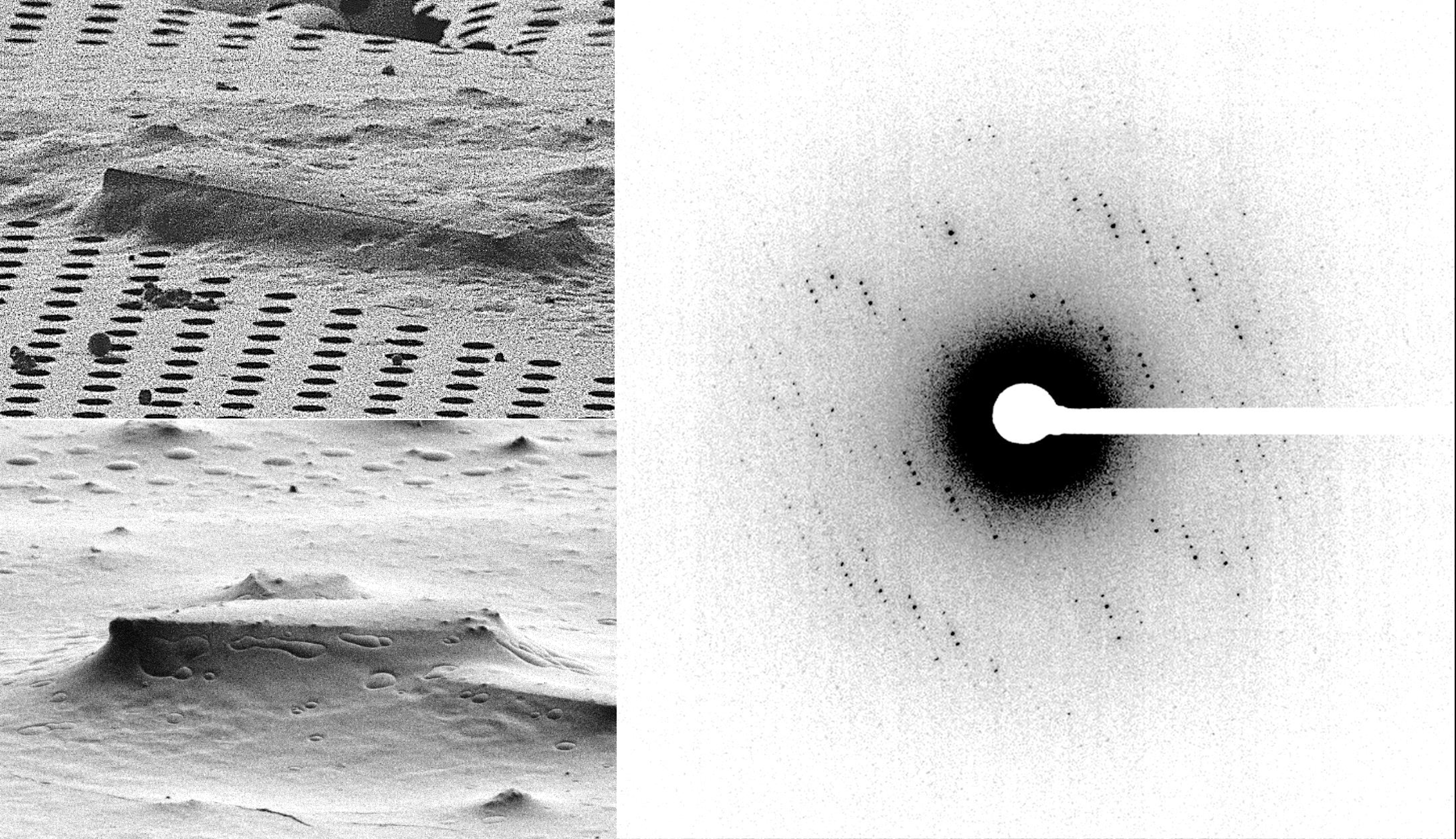
MicroED structure of the human adenosine receptor determined from a single nanocrystal in LCP.
Martynowycz MW, Shiriaeva A, Ge X, Hattne J, Nannenga BL, Cherezov V, and Gonen T.
Reviewed by: Young ID, Fraser JS.
Improvement of cryo-EM maps by density modification.
Terwilliger TC, Ludtke SJ, Read RJ, Adams PD, and Afonine PV.
Reviewed by: Young ID, Rohou A, Fraser JS.
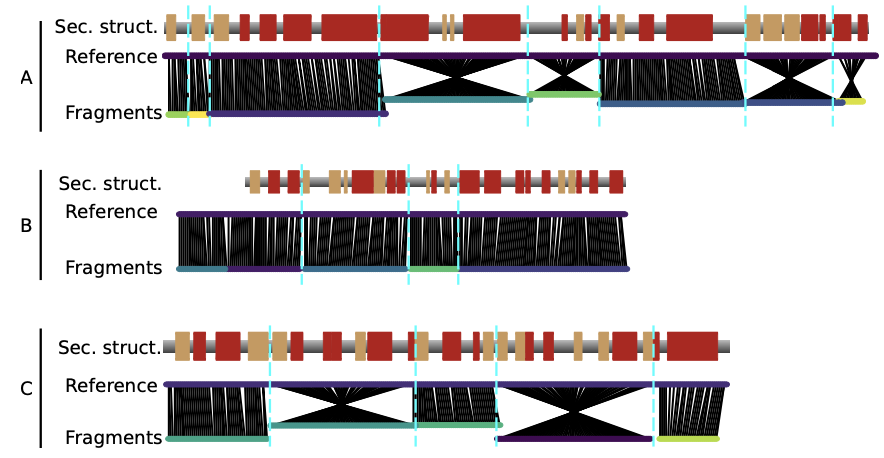
Automatic building of protein atomic models from cryo-EM density maps using residue co-evolution.
Bouvier G, Bardiaux B, Pellarin R, Rapisarda C, and Nilges M.
Reviewed by: Díaz RE, Young ID, Fraser JS.
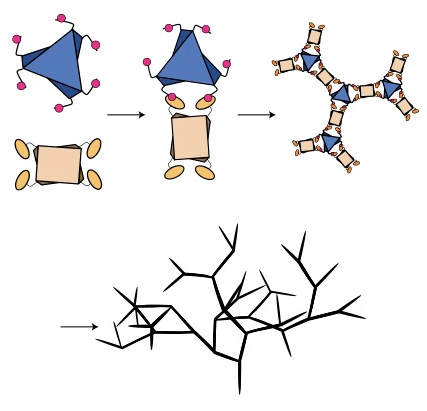
Stimulus-responsive self-assembly of protein-based fractals by computational design.
Hernández NE, Hansen WA, Zhu D, Shea ME, Khalid M, Manichev V, Putnins M, Chen M, Dodge AG, Yang L, Marrero-Berríos I, Banal M, Rechani P, Gustafsson T, Feldman LC, Lee SH, Wackett LP, Dai W, and Khare SD.
Reviewed by: Young ID, Fraser JS.
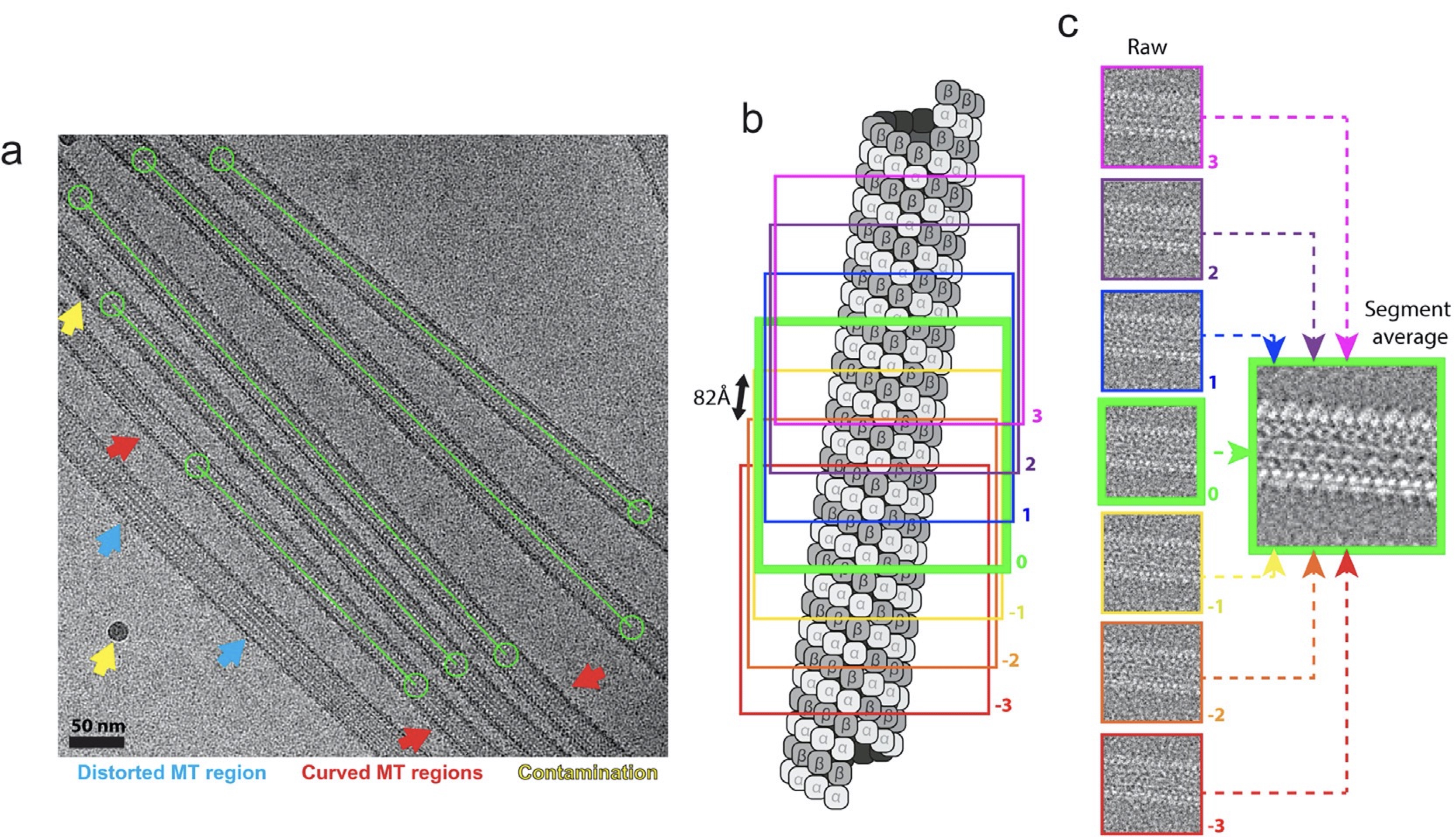
A microtubule RELION-based pipeline for cryo-EM image processing.
Cook AD, Manka SW, Wang S, Moores CA, and Atherton J.
Reviewed by: Young ID, Fraser JS.